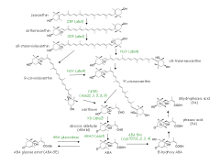
Abscisic acid (ABA)
Abscisic acid (ABA: (S)-(+)-2-cis-ABA) was originally identified as the bioactive compounds that induce abscission or bud dormancy. However, contribution of ABA to these physiological responses has not been clearly demonstrated. Instead, studies using ABA-deficient mutants from several plant species revealed that this hormone is required for the induction and maintenance of seed dormancy, stomatal closure, and response to biotic and abiotic stresses. (R)-(-)-2-cis-ABA (generally termed (-)-ABA) is biologically active as an ABA analogue, but do not exist in plant tissues. (S)-(+)-2-trans-ABA (trans-ABA) has been detected from plant tissues but do not exhibit ABA activity. ABA is synthesized via carotenoids. The biosynthesis pathway after zeaxanthin formation is shown here.
Genes on the pathway
| Gene name | Locus name | Description |
|---|---|---|
| ABA1/ZEP | At5g67030 | zeaxanthin epoxidase (ZEP) |
| ABA4 | At1g67080 | neoxanthin synthase (NSY) |
| NCED2 | At4g18350 | 9-cis-epoxycarotenoid dioxigenase (NCED) |
| NCED3 | At3g14440 | 9-cis-epoxycarotenoid dioxigenase (NCED) |
| NCED5 | At1g30100 | 9-cis-epoxycarotenoid dioxigenase (NCED) |
| NCED6 | At3g24220 | 9-cis-epoxycarotenoid dioxigenase (NCED) |
| NCED9 | At1g78390 | 9-cis-epoxycarotenoid dioxigenase (NCED) |
| ABA2 | At1g52340 | xanthoxin dehydrogenase (XD) |
| AAO3 | At2g27150 | abscisic aldehyde oxidase (ABAO) |
| ABA3 | At1g16540 | molybdenum cofactor sulfurare |
| CYP707A1 | At4g19230 | ABA 8’-hydorxylase (ABA8ox) |
| CYP707A2 | At2g29090 | ABA 8’-hydorxylase (ABA8ox) |
| CYP707A3 | At5g45340 | ABA 8’-hydorxylase (ABA8ox) |
| CYP707A4 | At3g19270 | ABA 8’-hydorxylase (ABA8ox) |
| BG1 | At1g52400 | ABA glucosidase |